Mar 29, 2019
Serverless extraction of large scale data from Elasticsearch to Apache Parquet files on S3 via Lambda Layers, Step Functions and further data analysis via AWS Athena
Feature extraction and dimensionality reduction are common and basic initial tasks in data preparation for machine learning. In most of our (Kubernetes) projects we use the common EFK stack (Elasticsearch + Fluentd + Kibana) which is good for ad-hoc troubleshooting but lacks large scale data analysis capabilities. To get to the bottom of bigger data sets a different approach is needed. Here I explore an approach in which extracted Elasticsearch data (25m records or more) is stored as columnar data files in S3 and further slicing and dicing, with fast response times, executed with AWS Athena.
Columnar data formats are key for BI and data analytics workloads. They help to optimize the analytics query performance, because they drastically reduce the overall disk I/O requirements, and also reduce the amount of data to load from disk. The most commonly used formats are Apache Parquet or Apache ORC. In this article I investigate how to extract Elasticsearch data on a large scale to do further analysis and extraction.

Table of Contents
- Serverless extraction of large scale data from Elasticsearch to Apache Parquet files on S3 via Lambda Layers, Step Functions and further data analysis via AWS Athena
- Elasticsearch bulk export via scrolling
- Execution as step function
- Data storage as columnar file
- Package all dependencies as Lambda Layer
- Data access via AWS Athena
- Conclusion
- Appendix
- Snappy Lambda runtime issues
- Fastparquet installation issues
Elasticsearch bulk export via scrolling
Elasticsearch has a http based API to retrieve large numbers of results from a search request, in much the same way as a cursor would be used on a traditional database. The inital call includes a scroll parameter in the query string, which tells Elasticsearch how long it should keep the “search context” alive.
from elasticsearch import Elasticsearch
es = Elasticsearch("http://myserver:13000/elasticsearch")
page = es.search(
index = myindex,
scroll = '2m',
size = 10000,
body = myquery)
sid = page['_scroll_id']
scroll_size = page['hits']['total']
The scroll parameter (passed to the search request and further on to every scroll request) tells Elasticsearch how long it should keep the search context alive and needs to be long enough to process the batch of results before the next scroll request. Now it is possible to iterate and retrieve the data associated to the query:
while (scroll_size > 0):print("Scrolling...", i)page = es.scroll(scroll_id = sid, scroll = '2m')# Update the scroll IDsid = page['_scroll_id']# Get the number of results that we returned in the last scrollscroll_size = len(page['hits']['hits'])print("scroll size: " + str(scroll_size))# Convert the page into a Pandas dataframedfES = Select.from_dict(page).to_pandas()dfES.drop(columns=['_id', '_index', '_score', '_type'], inplace=True)
It should be noted that the maximum value for size, the number of hits to be returned with each batch of results, is 10000. In my case it is a data set of 25m records which needs therefore 2500 scroll iterations of the batch extraction. To reduce the number of iterations, in a Lambda function 15 scroll events are executed during one invocation, which yields 150,000 records. Together with the further processing of the results, each Lambda invocation takes around 60 seconds, therefore the overall processing time is around 3 hours. (If the data set is even larger, it is possible to execute parallel a sliced scrolling on different nodes of the Elasticsearch cluster). The overall execution job runs as a Step Function with a single Lambda execution per extraction batch.
Execution as step function
To automatically execute the scrolling Lambda until the last page is extracted a Step Function is the perfect choice.
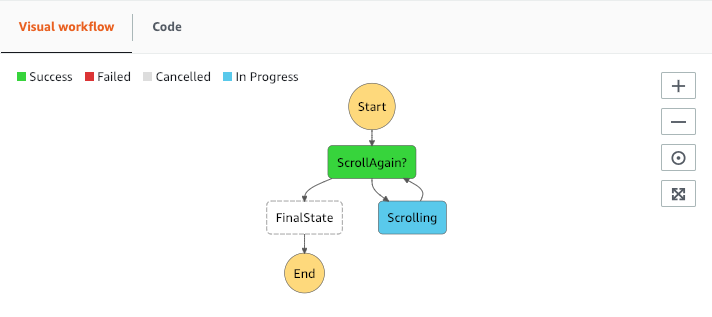
Here the output of the Lambda is checked and if it indicates that further pages are still available, the Lambda function is executed again. The Step function can be triggered by a Cloudwatch event, by indicating "state": “Start" , at a given time and orchestrates extraction of all data. The data/event passed between the runs is like:
{
"state": "Scroll",
"scrollId":
"DnF1ZXJ5VGhlbkZldGNoBQA2VjRfUUJRAAAAALckuFkY5bS1IbDhzUk5LUFlXMjZWNF9RQlEAAAAAAC3JLxZGOW0tSGw4c1JOS1BZVzI2VjRfUUJRAAAAAAAAApAWbGtqLUVOQWVSZm03NmdxTllqNFhZZwAAAAAALckxFkY5bS1IbDhzUk5LUFlXMjZWNF9RQlEAAAAAAC3JMBZGOW0tSGw4c1JOS1BZVzI",
"index": 12879232
}
The possible states are Start, Scroll and Done. The state function is defined as following:
{ "StartAt": "ScrollAgain?", "States": { "ScrollAgain?": { "Type" : "Choice", "Choices": [ { "Variable": "$.state", "StringEquals": "Done", "Next": "FinalState" } ], "Default": "Scrolling" },"Scrolling": { "Type" : "Task", "Resource": "arn:aws:lambda:eu-west-1:xxxxxxxxx:function:MyParquetfunction", "Next": "ScrollAgain?" },"FinalState": { "Type": "Succeed" } }}
Data storage as columnar file
The extracted data could be stored as a CSV file, as is typical in many analytics use cases, to allow further processing. However, since the dataset is quite large such an approach is not really a viable option.
Storing the data extract in several data files and specifically as a columnar-storage file has several advantages especially when using the data with Athena. Apache Parquet is column-oriented and designed to bring efficient columnar storage of data compared to row based files like CSV. It has features like dictionary encoding, bit packing and run-length encoding (RLE) to compress the data. This blog post “Dremel made simple with Parquet” from the Twitter engineering team gives a more detailed overview.
Additional compression can be applied to the parquet file, it is even possible to apply different compression algorithms per column.
Snappy is a light and fast compression codec and it does not need much CPU utilization, but it does not compress that well as gzip or bzip2. In this case applying snappy compression reduced the file size by a factor of 5, and it was 2 times bigger than the same file gzip compressed.
In Python it is quite easy to write a parquet file and integrate the upload to S3:
import s3fsimport fastparquet as fpimport pandas as pdimport numpy as nps3 = s3fs.S3FileSystem()myopen = s3.opens3bucket = 'mydata-aws-bucket/'# random dataframe for demodf = pd.DataFrame(np.random.randint(0,100,size=(100, 4)), columns=list('ABCD'))parqKey = s3bucket + "datafile" + ".parq.snappy"fp.write(parqKey, df ,compression='SNAPPY', open_with=myopen)
This example will generated and upload a Snappy compressed parquet file. In the actual use case groups of 10 pages were combined in one parquet file of around 40MB in size and the extraction of the 25m events from Elasticsearch generated around 200 of these files, in total a 6GB data set.
Pandas and Numpy should be well known as standard Python data science libraries. Fastparquet is an interface to the Parquet file format that uses the Numba Python-to-LLVM compiler for speed. It is a fork by the Dask project from the original implementation of python-parquet by Joe Crobak. S3Fs is a Pythonic file interface to S3 which builds on top of boto3.
Package all dependencies as Lambda Layer
Packaging the dependencies to be uploaded is along the line of my last blog post “Pure serverless machine learning inference with AWS Lambda and Layers”via a zipped package. See below that the LD_LIBRARY_PATH has to be set accordingly for the Lambda function to access the compiled Snappy library within the layer. Also building with Pip and an additional target value caused some issues with the fastparquet installation, more details are provided below.
rm -rf python && mkdir -p pythondocker run --rm -v $(pwd):/foo -w /foo lambci/lambda:build-python3.7 /foo/build.sh
The “external” target directory is cleaned and then a build matching the AWS runtime is executed within the container. The build.sh looks like following:
#!/usr/bin/env bash
export PYTHONPATH=$PYTHONPATH:/foo/python
yum install snappy-devel -y
pip install --upgrade pip
pip install -r requirements.txt --no-binary --no-dependencies --no-cache-dir -t python
pip install fastparquet==0.2.1 --no-binary --no-dependencies --no-cache-dir -t python
cp /usr/lib64/libsnappy* /foo/lib64/
As said, the fastparquet module has to be manually removed from the requirements.txt file and installed as a second step due to the usage of the -t python target parameter. Then the layer can be deployed like:
zip -r MyParquet_layer.zip python lib64aws s3 cp MyParquet_layer.zip s3://mybucket/layers/aws lambda publish-layer-version --layer-name MyParquet_layer --content S3Bucket=mybucket,S3Key=layers/MyParquet_layer.zip --compatible-runtimes python3.7
Data access via AWS Athena
To further work with the data and extract some subsets, AWS Athena is the perfect tool. AWS Athena is a serverless interactive query service that makes it easy to analyze large amounts of data in S3 using standard SQL. It handles columnar and compressed data like Parquet or ORC with Snappy out of the box.
An Athena table for the data in S3 can easy be created via the command:
CREATE EXTERNAL TABLE IF NOT EXISTS myanalytics_parquet (`column1` string, `column2` int, `column3` DOUBLE, `column4` int, `column5` string )STORED AS PARQUETLOCATION 's3://mybucket/parquetdata/'tblproperties ("parquet.compress"="SNAPPY")
After this the data is already available and can be queried with an incredible speed and response time.
SELECT column5, count(*) AS cnt FROM myanalytics_parquetGROUP BY column5ORDER BY cnt DESC;
A query like this takes only a few seconds to execute on the complete data set. The resulting data is available in the default S3 bucket assigned the Athena instance and can be extracted as CSV files.
Conclusion
Extracting data in batches from Elasticsearch and converting it into compressed Parquet files which are stored in S3 is an effective approach to work with large data sets. Using AWS Athena with these files makes it easy to slice and dice the dataset in a fast and efficient way and allows get a defined subset of data (dimensionality reduction) out for further processing or feature extraction. It is for sure a faster and more affective way than working with the original data set in Elasticsearch. Getting data out of Elasticsearch can be done via the scroll function, and running this with a step function and a lambda is an effective serverless solution.
Appendix
Installing snappy and fastparquet into a lambda function is not so easy, since they both use C-extensions and libraries.
Snappy Lambda runtime issues
Regarding the snappy installation I ran into following issue:
ModuleNotFoundError: No module named 'snappy._snappy_cffi'
Unfortunately this was a masked error, an initial exception occurred by not finding the libsnappy.so library and then an additional exception occurred as seen above, which initially led me down the wrong track. The issue here, is that Lambda Layers are all mapped under the /opt directory . This issue report libraries under /opt are not detected #12 in the aws-lambda-container-image-converter repository gave the clue that the LD_LIBRARY_PATH had to be extended and set for the Lambda function like:
$LAMBDA_TASK_ROOT/lib:$LAMBDA_TASK_ROOT/lib64:$LAMBDA_RUNTIME_DIR:$LAMBDA_RUNTIME_DIR/lib:$LAMBDA_TASK_ROOT:/opt/lib:/opt/lib64:/lib64:/usr/lib64
and then place the shared library in the lib64 folder, where it will be found.
Fastparquet installation issues
The fastparquet installation also caused some trouble. If the Pip command is executed with a target parameter, the fastparquet build cannot find the necessary numpy header files. One way fixing this is to remove fastparquet from the requirements.txt file and install it as a second step. Then the numpy headers can be found when the PYTHONPATH is set correctly.